What does PCR stand for?
Polymerase chain reaction
Which nonsynonymous mutation changes a codon to a stop codon?
Nonsense
What is the name of the step that turns DNA into pre-mRNA and what synthesizes RNA from DNA?
Transcription
RNA polymerase
What allows the GFP gene to glow?
sugar (arabinose)
What is the genome?
organism complete genetic information
What’s DNA’s charge and which way does it travel?
Negative (phospates), positive
cDNA are shorter than gDNA…WHY?
reverse transcript of RNA doesn't include introns; splicing
Name one stop codon
UAA/UGA/UAG
U Are Annoying/U Go Away/U Are Gone :,(
Name the 2 types of promoters and the genes that we used for Experiment 3:
Constitutive: Amp-R
Conditional: GFP
What is a transcription factor?
Proteins that bind to DNA and regulate gene expression
If you dilute a solution that has a volume of 300 mL and a concentration of 90 M to make a solution that has a final volume of 500 mL, what will the final concentration be and how much water will you have added?
final concentration = 54M
water volume = 200mL
c1v1 = c2v2
90M * 300mL = c2 * 500mL
final volume =(90*300)/500 = 54M
volume of water = final volume - initial volume
500 - 300 = 200mL
Explain difference between Missense Synonymous, Missense nonsynonymous, and Nonsense.
Synonymous: same amino acid
Nonsynonymous: different amino acid
Nonsense: stop
This is the term for a three-nucleotide sequence on mRNA that codes for a specific amino acid.
codon
How can a bacteria uptake plasmid?
Bacteria is turned chemically competent by weakening their cell walls through chemical washes. This allows plasmids to enter after co-incubation with bacteria on ice followed by heat shock.
Name one way to identify genetic differences in organisms
Genomic sequencing
What step of PCR involves the separation of DNA strands and what temperature does it require. What is the enzyme involved in PCR?
Denaturation/ 98 C/ DNA polymerase
The mRNA sequence "AUC UAC AGC GAA UAG" codes for the amino acids "I-T-S-G-Stop". The third codon has a SNP mutation that changes the third nucleotide 'C' to an 'A': AGC --> AGA (changing the protein to A) What type of mutation would this be?
Nonsynonymous -missense
What type of RNA serves as a link for growing amino acid chain?
Transfer-RNA/tRNA
Why is GFP conditional?
The GFP promoter is conditional because of a transcription factor (regulates gene expression).
In the case of GFP, there is a repressor that blocks transcription of the GFP gene.
However, arabinose binds to this repressor, changing its shape and causing it to be unable to bind to the promoter anymore. This frees up the promoter, allowing RNA polymerase to bind and create mRNA for expression.
Transcription vs Translation
Transcription:
Nucleus.
DNA serves as a template to produce mRNA.
RNA polymerase: binds to the DNA in its promoter region and turns it into pre-mRNA (both Exons and introns)
After transcription: pre-mRNA gets spliced by the spliceosome → turns into mRNA
Translation:
mRNA to Cytoplasm/ goes through the ribosome
The ribosome reads the mRNA nucleotide sequence by 3 (codon) with the help of rRNA
Transfer RNA (tRNA) carry anti-codons to the ribosome → corresponds to particular amino acid
Forms a polypeptide chain till stop codon. (UAA, UAG, UGA)
What are the 3 steps of PCR (and what do you do in them)? What is PCR purpose?
Amplify certain gene/region in a DNA
3 steps of PCR: DAE
Denaturation:
Heat breaks the hydrogen bonds in a DNA to separate the strands
Annealing:
Primers bind to single stranded DNA
Remember: primers are short DNA used to identify/stick to the specific gene in a DNA to amplify it
Extension:
DNA polymerase go through the single strands of the marked DNA and turn in into double strand
Using PCR & gel electrophoresis, which template and primers could you use to detect a SNP in Exon 1?
Option 1) gDNA, primers L1 and R1
Option 2) cDNA, primers L2 and R2
Neither of these options would work
Both of these options would work
Neither of these options would work
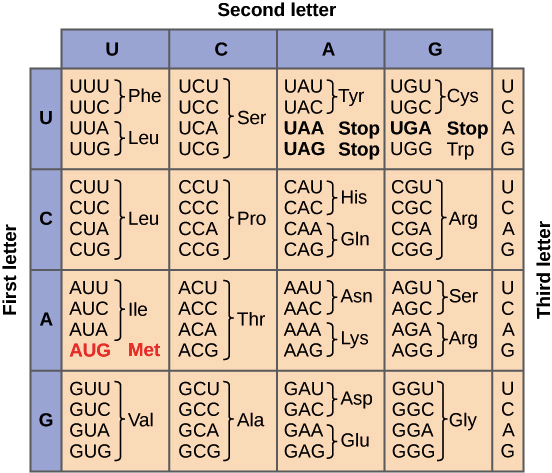
Using the codon table, look at the following mRNA sequence (ACG AAA UCU CAC UAA), what would the correct string of amino acids be if the second nucleotide "C" of the third codon was deleted?
Thr Lys Phe Thr----
Transformation efficiency formula?
first calculate mass of plasmid: #ng of plasmid = volume of DNA used * concentration of DNA.
transformation efficiency: (# colonies/ #ng of plasmid) * (#total volume µl/#plated volume)