What are the three main "parts" of a nucleotide? Sketch and label them.
Nitrogenous base, phosphate group, sugar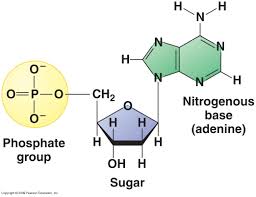
List 3 similarities and 3 differences between DNA polymerase and RNA polymerase.
Similarities between DNA polymerase and RNA polymerase:
1) Both DNA polymerase and RNA polymerase acts on DNA for replication and transcription respectively.
2) Both the enzymes perform polymerisation by adding nucleotide units to each other.
3) Both the enzymes are present in nucleus of a eukaryotic cell.
Differences between DNA polymerase and RNA polymerase:
1) DNA polymerase is used in replication of DNA while RNA polymerase is used in formation of RNA from DNA in the process of transcription.
2) DNA polymerase synthesizes a double standed DNA on replication while RNA polymerase synthesizes a single standed RNA on transcription.
3) DNA polymerase requires a primer to start the process of replication while RNA polymerase does not require any primer for transcription process.
Explain the following terms and say which molecule they are found:
gene
promoter
enhancer
gene: a distinct sequence of nucleotides forming part of a chromosome, the order of which determines the order of monomers in a polypeptide or nucleic acid molecule which a cell (or virus) may synthesize.
promoter: DNA sequences that defines where transcription of a gene by RNA polymerase begins
enhancer: enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur
What are two limitations on DNA polymerase activity?
It can’t start strands, it needs a primer
It can’t add new Nucleotides to the 5’ end of growing DNA chains, which causes okazaki fragments
Explain what energy source drives the polymerization of DNA.
Comes from 2 of the 3 phosphates attached to each unincorporated base. When a nucleotide is being added to a growing DNA strand, 2 of the 3 phosphates are removed and the energy produced creates a phosphodiester bond that attaches the remaining phosphate to the chain.
Why are DNA strands antiparallel and why is it important?
The nucleic acid sequences are complementary and parallel, but they go in opposite directions, hence the antiparallel designation. The antiparallel structure of DNA is important in DNA replication because it replicates the leading strand one way and the lagging strand the other way
Explain (in words or pictures) the steps to translation.
initiation: the ribosome assembles around the mRNA to be read and the first tRNA
Elongation: the amino acid chain gets longer. In elongation, the mRNA is read one codon at a time, and the amino acid matching each codon is added to a growing protein chain
Termination: a stop codon (UAG, UAA, or UGA) enters the ribosome, triggering a series of events that separate the chain from its tRNA and allow it to drift out of the ribosome
What are 2 major differences between prokaryotic and eukaryotic gene regulation?
Eukaryotic genes are NOT regulated by operons.
In eukaryotes transcription and translation are separated by the nuclear membrane
How does semi-conservative replication help prevent mutations during DNA replication?
one half of the original molecule is kept and the new strand is made from free nucleotides, which can only add according to the complementary base-pair rule, so it lessens the likelihood that the wrong nucleotide sequence is created based on the original
Explain why continuous synthesis of both DNA strands is not possible.
The leading strand is able to continuously synthesize DNA. However, the lagging strand is not able to. This is bc it, while synthesizing in the 5' to 3' direction, would run into previously synthesized DNA. Other primers will go in further behind the synthesized DNA to allow more to DNA Polymerase to synthesize more DNA. This creates Okazaki fragments which are bonded together by DNA ligase.
What does the complementary base-pair rule state?
adenine and thymine form pairs, and cytosine and guanine form pairs
Draw/describe the structure of the ribosome complexed with mRNA at the start codon. Label the A,P,E sites.

Explain how an activator and an repressor works.
activator: a protein (transcription factor) that increases gene transcription of a gene or set of genes. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements.
repressor: can turn on/off an operon. binds to operator & blocks attachment of RNA pol to promoter
List all the enzymes used in DNA replication and describe their functions.
Helicase: unwinds parental double helix at replication forks
Single-Stranded Binding Protein: binds to and stabilizes single-stranded DNA until it can be used as a template
Topoisomerase: Relieves "overwinding" strain ahead of replication fork by breaking, swiveling, and rejoining DNA strands
Primase: Synthesizes an RNA primer at 5' end of leading strand of each okazaki fragment of the lagging strand
DNA Pol III: Using parental DNA as a template, synthesizes a new DNA strand by covalently adding nucleotides to the 3' end of a pre-existing DNA strand or RNA primer
DNA Pol I: Removes ENA nucleotides of primer from 5' end and replace them with DNA nucleotides
DNA Ligase: joins 3' end of DNA that replaces primer to rest of leading strand and joins okazaki fragments of lagging strand
Explain how RNA polymerase recognizes where transcription should begin.
RNA polymerase recognizes the promoter of nucleotides (AUG).
Give the complimentary DNA strand from this RNA:
AGGCGUUAGA
TCCGCAATCT
Translate this mRNA sequence into proteins.
AUGGUGUCACGGGGGGAUUAG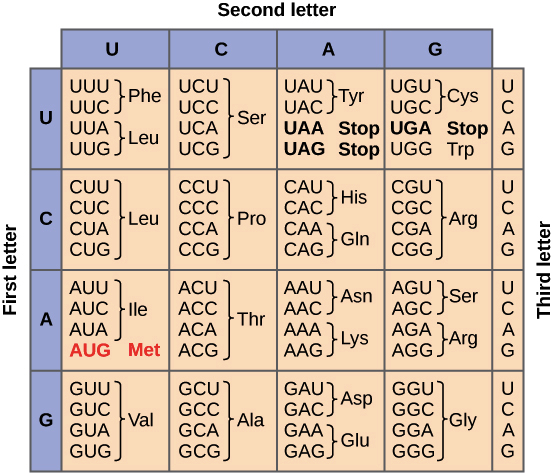
Met Val Ser Arg Gly Asp STOP
When DNA is _______, it stays compressed and is unavailable for transcription.
a. phosphorylated
b. methylated
c. acetylated
d. degraded
b. methylated
Draw and label a simple diagram of DNA replication.

Describe the three general post transcriptional modifications made to eukaryotic RNA in the nucleus and why they are necessary.
Addition of 5' cap (modified Guanine nucleotide)
Addition of 50-250 adenines at the 3' end (poly-A tail)
Removal of noncoding internal sequences from RNA (introns)
What is the template and the coding strand?
Template strand: the strand used by DNA polymerase or RNA polymerase to attach complementary bases during DNA replication or RNA transcription
Coding Strand: doesn't really do anything; the other strand
Compare and contrast transcription and translation in prokaryotes and eukaryotes.
Transcription: DNA -> RNA
Translation: RNA -> Protein
What is the difference between basal transcription factors and regulatory (specific) transcription factors?
general transcription factors (basal) are required by all mRNA genes (you can use only general TFs to get the basal level transcription) , but promoter-specific TFs are different for each gene (can reach maximal level of transcription)
Summarize the steps of DNA replication.
1. hydrogen bonds between nucleotides break
2.strands of DNA separate
3. free nucleotides are attracted to exposed bases on the loose strands of DNA
4. hydrogen bonds between nucleotides form
Describe the concepts of positive and negative regulation of operon transcription.
Negatively regulated: genes normally on, and binding a repressor protein to the DNA turns transcription off
Positively regulated: genes are normally off, and binding an activator protein to the DNA turns transcription on