What is the first enzyme in DNA replication
Topoisomerase
What is the role of primase in DNA replication?
Sets RNA primer (Okazaki fragments) for DNA polymerase III to build off of.
What are 3 errors in DNA replication that can lead to a small scale mutation?
1. Substitution (nucleotide replaced)
2. Deletion (removal of nucleotide)
3. Insertion (addition of nucleotide)
Bonus (answer in 200pt column): What are the possible effects of these small scale mutations? 50pts for each one given.
Why is DNA considered a polymer and give monomer name
It is made up of monomers called nucleotides
What are the 4 nitrogen bases and which 2 are considered Purines and which 2 are Pyrimidines. (100pts for full answer).
Adenine and Guanine are Purines and have double ring structure
Cytosine and Thymine are pyrimidines and have single ring structure.
Place these enzymes in order of when they occur in DNA replication.
Topoisomerase, SSBPs, helicase, DNA polymerase I, DNA polymerase III, primase, ligase
Topoisomerase, helicase, SSBPs, primase, DNA polymerase III, DNA polymerase I, ligase
What is the purpose for having multiple origins of replication in DNA replication.
3 Similarities or differences between DNA replication and transcription in protein synthesis
e.g
1. DNA replication creates exact copy of itself transcription copies info into RNA message
2. Both occur in nucleus
3. DNA replication replicates both strands, transcription only transcribes complimentary strand to the template strand
4. Different enzymes, DNA polymerase vs. RNA polymerase
Bonus (500 pt column) - Why can't proteins be directly synthesized from DNA? 200pts
The 3 parts making up nucleotides
1. Deoxyribose sugar
2. Phosphate group
3. Nitrogenous base
Bonus (answer in 100pt column): What are the 4 nitrogen bases and which 2 are considered Purines and which 2 are Pyrimidines. (100pts for full answer).
What are the possible effects of these small scale mutations? 50pts for each one given.
1. Silent mutation - nucleotide is replaced but the codon still produces the same amino acid
2. Missense - nucleotide is replaced and the codon now results in a different amino acid
3. Nonsense - The codon now result in stop command ending the protein at the location where the mutation occurred.
What are the 3 stages of replication
1. Strand seperation
2. Strand replication
3. Proof reading
Bonus (answer in 300pt column) - Explain what happens in each stage. 100pts for each correct explanation
What must be built on the lagging strand to allow for replication to occur that is not needed on the leading strand
RNA primer
These enzymes proofread during DNA replication to avoid mistakes or mutations
DNA polymerase I and III check all base pairings throughout DNA replication.
What direction is the leading strand synthesized in? (towards or away from the replication fork?)
Towards replication fork
Explain what happens in each stage of DNA replication. 100pts for each correct explanation
1. Strand seperation - topoisomerase relieves tension in the double helix, so that helicase can break the bonds between the complimentary bases. SSBPs prevent re-annealing
2. Strand replication- DNA polymerase adds bases to 3' end of new strand. For leading strand this occurs continuously toward replication fork. For lagging strand occurs in fragments starting on RNA primer. RNA primer replaced with DNA bases by DNA polymerase I and ligase joins fragments together with phosphodiester bonds.
3. Proof reading- DNA polymerase I and III continuously check all base pairings throughout DNA replication
Order the Okazaki fragments 1,2,3 in which way they were created and explain why they are formed in this order. (ignore blanks)

Fragment one will be the furthest from the replication fork, 2 will be in the middle and 3 will be the closest to the replication fork. This occurs because DNA polymerase 3 cannot synthesize the strand in one go so it has to start at the origin and work its way in fragments backwards towards the replication fork.
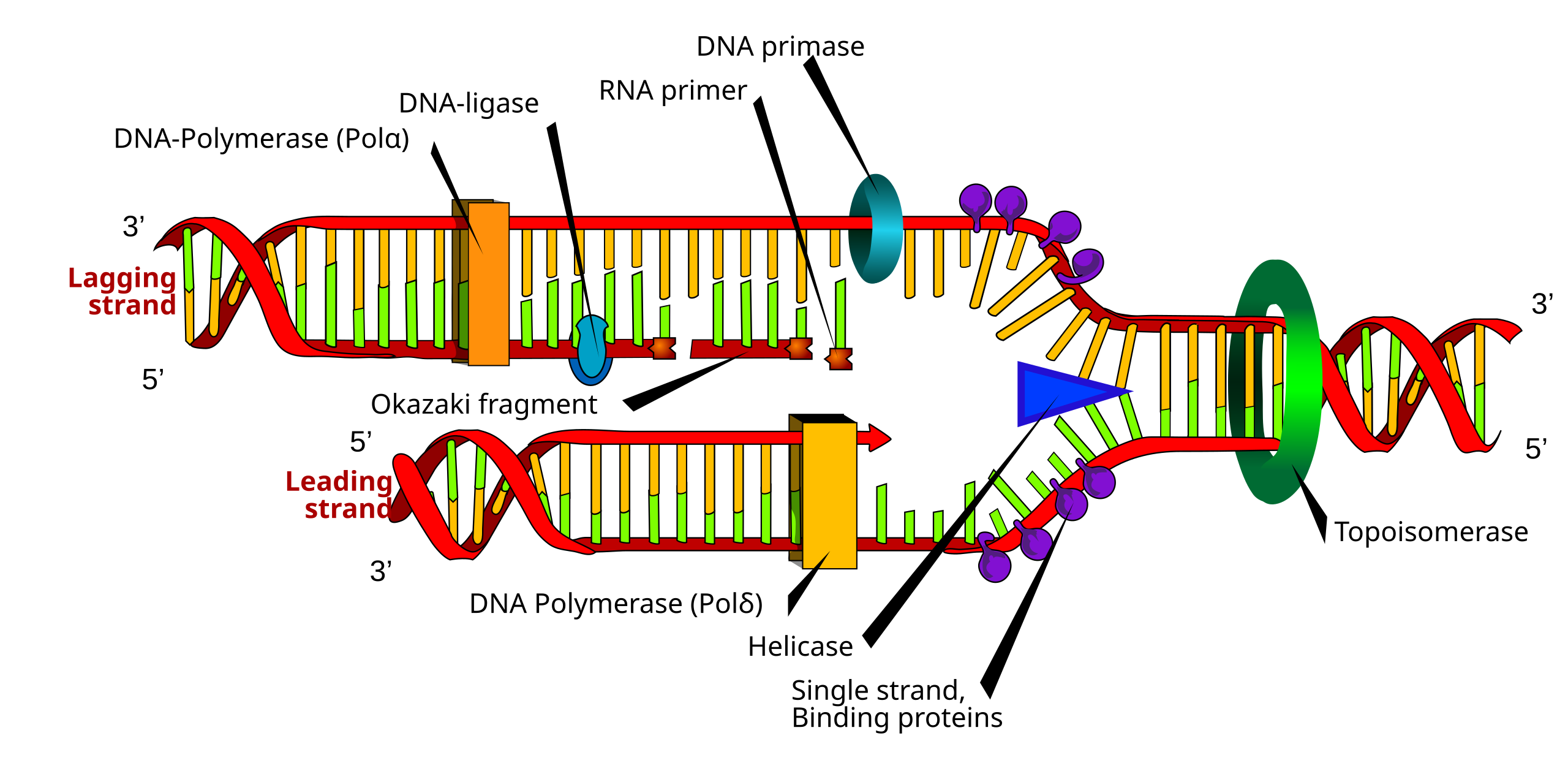
Fill in the blanks for 50 pts each

Note: DNA polymerase on lagging strand is DNA polymerase I and on leading strand is DNA polymerase III
Why are some mutations harmless or silent while others create large changes or termination of protein?
Some are harmless because nucleotide that got replaced in DNA replication does not change to amino acid produced. Others can be harmful because the codon then results in a different amino acid or even a stop codon which would terminate the protein all together.
Bonus (Answer is 400pt column) - Use your amino acid wheel to give an example of a 9 base long DNA sequence and alter it so that it would result in either a silent, missense or a nonsense mutation. 200pts
Why can both the leading strand and lagging strand not be replicated continuously?
The lagging strand cannot be replicated continuously because DNA polymerase III can only add complimentary base pairs in the 5' to 3' direction and because of the antiparallel nature of DNA it would have to go 3' to 5' direction and there is not an enzyme that can do that
Use your amino acid wheel to give an example of a 9 base long DNA sequence and alter it so that it would result in either a silent, missense or a nonsense mutation (multiple right answers). 300pts
e.g original strand - TAC ATG ACT... mRNA - AUG UAC UGA... Amino acids - Met Tyr stop
Silent mutation - TAC ATA ACT...
mRNA - AUG UAU UGA...
Amino acids - Met Tyr stop
missense mutation - TAC AGG ACT...
mRNA - AUG UCC UGA...
Amino acids - Met Ser stop
Nonsense mutation - TAC ATC ACT...
mRNA - AUG UAG UGA...
Amino acids - Met stop
How do these 4 enzymes work together to create lagging strand in DNA replication?
DNA polymerase I, Primase, DNA polymerase III, and Ligase.
Primase sets the RNA primers down to act as a starting place for DNA polymerase III to work off of. DNA polymerase I removes RNA nucleotides and adds equivalent DNA nucleotides. Lastly ligase fills in the gaps between the fragments.
Describe 3 roles of DNA polymerases in replication
1) Adding complimentary bases to template strand
2) Changing RNA to DNA
3) Proofreading to avoid mistakes in DNA replication
What are the 6 large scale mutations (50pts each given) and how are large scale mutations different from small scale?
Large scale is different from small scale because large scale affects entire portions of the chromosome whereas small scale affects DNA at a molecular level, changing the sequence of nucleotides.
1) Deletion (the loss of one or more genes from the parent chromosome)
2) Duplication (addition of one or more genes that are already present in the chromosome)
3) Inversion (reversal of one or more genes)
4) Insertion (gene(s) removed from one chromosome and added to another)
5) Translocation (Chromosomes swap one or more genes with another chromosome)
6) Non- disjunction (chromosomes not separated properly resulting in missing or extra chromosomes)
What would happen if these 2 enzymes were not coded for or available for DNA replication and how would it affect the process (250pts for each enzyme).
1) Ligase
2) Primase
1) If ligase was not available then the Okazaki fragments would not be joined together and there would be gaps in any newly created DNA which could cause it to break.
2) If primase were not available DNA polymerase would have no place to start as the RNA primers could not be build. This would result in DNA replication not happening at all
Bonus (500 pt column) - Why can't proteins be directly synthesized from DNA? 200pts
Protein synthesis occurs in the cytoplasm and DNA is kept in the nucleus and is too large to be transported out. Instead, it is transcribed onto mRNA to be transported out of the nucleus and used as a blueprint. This also protects the DNA from damage.