Why do we conduct PCR in an experiment?
to amplify a tiny amount of DNA to make it easier to analyze
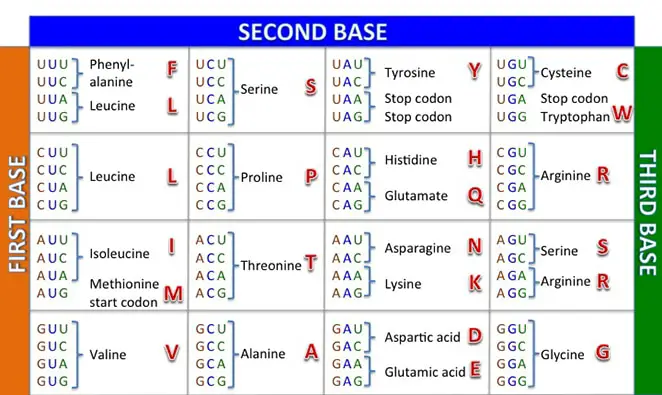 Translate the following mRNA sequence into the
Translate the following mRNA sequence into the
correct string of amino acids by the one letter
abbreviation: AUG UGG GAU UAC UAG
M-W-D-Y-Stop
M-W-E-D-Y
M-L-V-E-Stop
M-V-D-A-Y
M-Y-H-A-T
M-V-D-Y-Stop
M-W-D-Y-Stop
This type of RNA carries the genetic code from DNA and serves as a template for protein synthesis during translation.
mRNa --> messenger- RNA
True or False
Any type of gene can glow in the presence of Arabinose
False
A signaling molecule in smoke that is responsible for triggering germination
Karrikins
What step of the PCR reaction involves primers binding to the DNA template strands? (and at what temperature?)
Denaturation
Attaching
Extension
Annealing
Annealing 52/54
Gene editing is used to delete 100 bp in Intron 3 to
make it 50 bp long, which primers and template could
be used to detect this deletion?(Select all that apply).
gDNA with primers L2 and R2
cDNA with primers L1 and R2
cDNA with primers L2 and R2
gDNA with primers L2 and R1
gDNA with primers L2 and R2
Which of the following statements about transcription is incorrect?
Transcription is one of the steps required for genes to be ultimately expressed as protein.
DNA polymerase transcribes gDNA into pre-mRNA.
The end product of transcription is pre-mRNA.
pre-mRNA must be spliced to produce mRNA.
DNA polymerase transcribes gDNA into pre-mRNA
What antibiotic was used in our project to help make colonies?
Gentamicin
What do F-box proteins do?
targets proteins that are not needed and marks them for destruction
Which of the following statements about setting up a PCR reaction is incorrect? (2 correct answers)
1) Template for all samples should be added to the master mix before aliquoting into strip tubes.
2) The C1V1=C2V2 equation should be used to determine what volume of MyTaq is needed for the reaction.
3) Water can be treated as a sample to create negative control.
4) After all components are added to create the mastermix it should not be mixed.
1 and 4
The mRNA sequence "AUG GAA CGA UAC UAG"
codes for the amino acids "M-E-R-Y-Stop". The third
codon has a SNP mutation that changes the first
nucleotide 'C' to an 'A': What type of mutation would
this be?

Nonsense nonsynonymous
Missense nonsynonymous
Synonymous
Transposable element
Synonymous does not change the protein
What removes introns?
Splicosome
Define bacterial transformation
process allowing bacteria to acquire new genetic material by taking up naked DNA from their surroundings
Rank the following metric system from largest to smallest
nano, 1, kilo, pico, micro, milli
kilo
1
milli
micro
nano
pico
How much agarose is required to make a 3% gel with a volume of 300 mL 1X TAE?[Hint: 1% gel = 0.5g in 50 mL TAE]
0.3 g
9 g
3 g
0.9 g
3/100 * 300mL = 9 g
The mRNA sequence "AUG UUC ACC GAA UAC" codes for the amino acids "M-F-T-E-Y". What nucleotide in the fifth codon would you change to introduce a nonsense mutation and to what?
UAC to UAA or UAG
Which molecule transports amino acids to the ribosome, where they are added to the growing polypeptide chain during translation.
T-RNA
What is the name of the type of cells that were used during the Promoter Activation Experiment that allowed them to take up the plasmid DNA easier?
Competent
Constitutive
Complacent
Conditional
Capable
Competent
If you dilute a solution that has a volume of 210 mL and a concentration of 75 M to make a solution that has a final volume of 525 mL, what will the final concentration be and how much water will you have added?
30M and 315mL of water
Using the mRNA sequence (AUG UGG GAU UAC UAG), what would the correct string of amino acids be if the first nucleotide "U" of the second codon was DELETED?

M-G-I-T-... ("AG" nucleotides leftover)
M-D-Y-Stop
M-V-E-Stop
M-G-D-L-... ("G" nucleotide leftover)
M-W-D-Y-... ("AG" nucleotides leftover)
M-G-I-T-... ("AG" nucleotides leftover)
Where do transcription and translation occur in cell?
Transcription: nucleus
Translation: cytoplasm
You are performing a bacterial transformation experiment using E. coli and a plasmid containing an antibiotic resistance gene. You added 10 µL of plasmid DNA (concentration = 200 ng/µL) to 100 µL of E. coli cells. After performing the transformation and plating the mixture on selective media containing antibiotic, you obtain 150 colonies on the plate. You plated 50 µL of the transformation mixture onto the agar plate.
Calculate the transformation efficiency (in colony-forming units per nanogram of plasmid DNA).
#ng of plasmid = volume of DNA used * concentration of DNA.
transformation efficiency: (# colonies/ #ng of plasmid) * (#total volume µl/#plated volume)
Mass of plasmid=10μL×200ng/µL=2000ng
Transformation Efficiency=(150/2000)×(100/50) = 0.15CFU/ng of plasmid
what was the overall goal of your project?
to determine which specific amino acids in the ACR5 protein are responsible for protein-protein interaction by introducing mutations into the ACR5 gene and how that would later effect germination