Describe the central dogma that applies to both prokaryotic and eukaryotic cells
Central Dogma is the principle that genetic information flows from DNA to RNA and then that RNA becomes a protein
DNA ----->>>> RNA------>>>> Protein
Describe the three RNA Polymerases in eukaryotic cells, and which one is the most heavily involved with transcription
RNA Polymerase I - transcribes rRNA genes
RNA Polymerase II - transcribed ALL protein coding genes so most involved, and some micro RNA genes, and some small nuclear RNA genes
RNA Polymerase III - transcribe tRNA genes, and some other numerous small functional RNA genes, and 1 type of rRNA
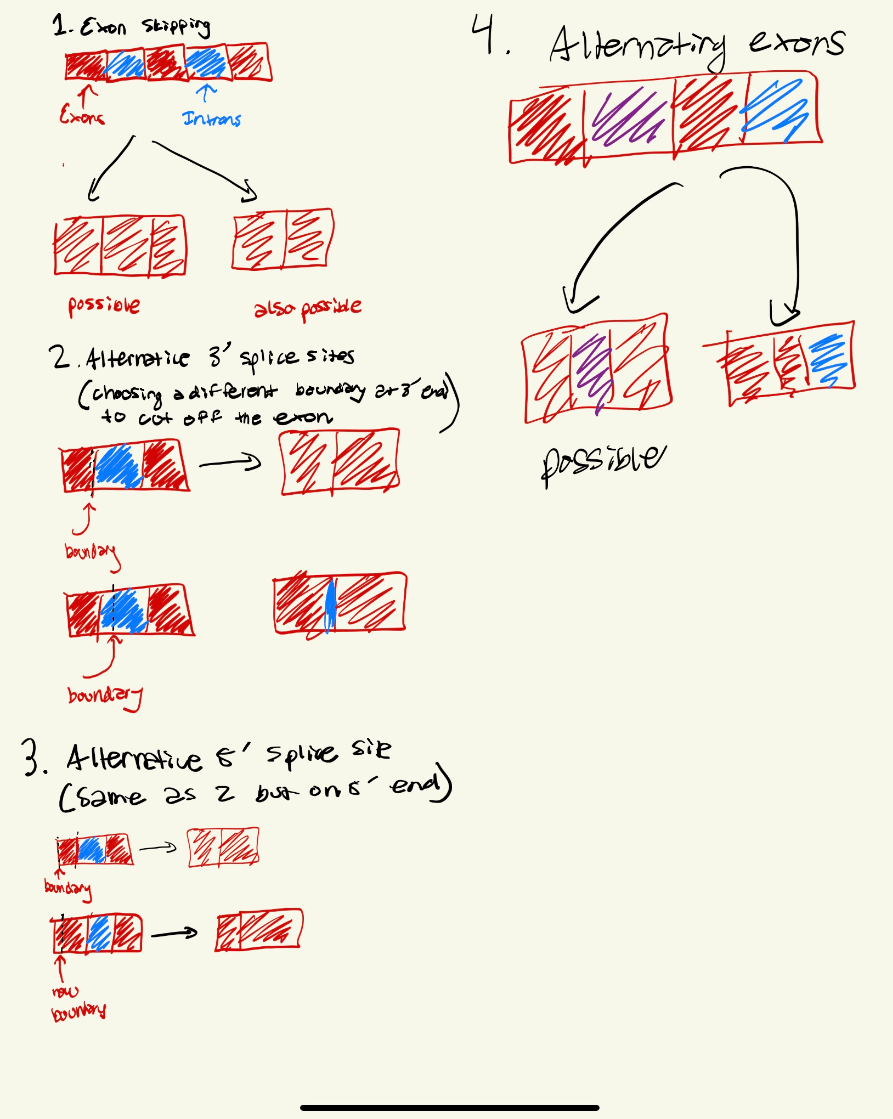
Alternative splicing allows for a single gene to code for multiple different proteins by varying combinations of exons in the final mRNA transcript, give examples of how this can occur.
Which of the answer choices would have the higher boiling point?
a. DNA strand made of 20% Adenine
b. DNA strand made of 15% Guanine
c. DNA strand made of 18 % Cytosine
d. DNA strand made of 9% Thymine
d.
9% T+9 % A=18% so remaining 82% is Guanine/Cytosine pairs, which have 3 h bonds instead of 2, so majority is G/C pairs, so d would be harder to melt.
Below is DNA being actively replicated, the replication fork in moving towards the right. What are the next two nucleotides that will be synthesized in the LAGGING strand?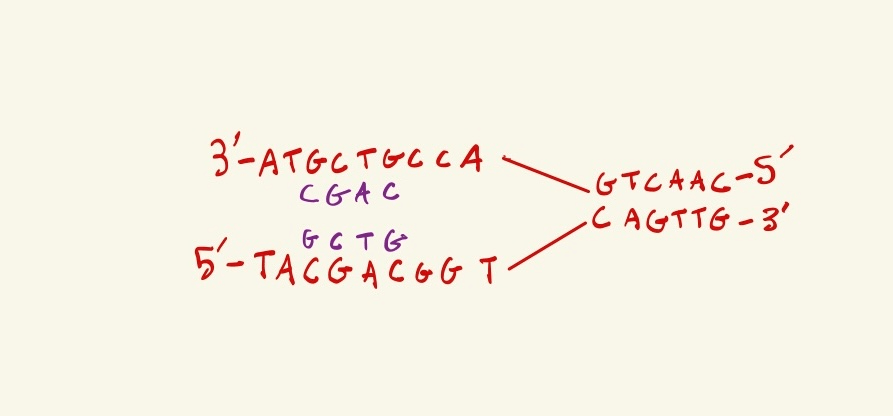
TA
1. Label the 5' and 3' ends of the new DNA strands being created
2. You find that the bottom strand is lagging, because the new DNA strand created from the bottom template strand can only be synthesized in the 5' to 3' direction, so it goes away from the replication fork, thus the new
Why is easier for the RNA Polymerase to access DNA in a prokaryote than a eukaryote?
The DNA of prokaryotes is not packaged into chromatin like eukaryotic DNA is.
Chromatin is a coiled complex of DNA wrapped around histone proteins along with other types of proteins as well
What has to be assembled at the promoter before RNA Polymerase comes?
GTF - General transcription factor
As rna polymerase comes, it brings MORE GTFs with it, but some have to be present already at promoter site.
1. Which sections represent exons?
2. Which sections are the introns
3. Where would the TATA binding protein bind?
4. Which sections would interact with the spliceosome complex?
5. Where would find the Poly (A) signal?
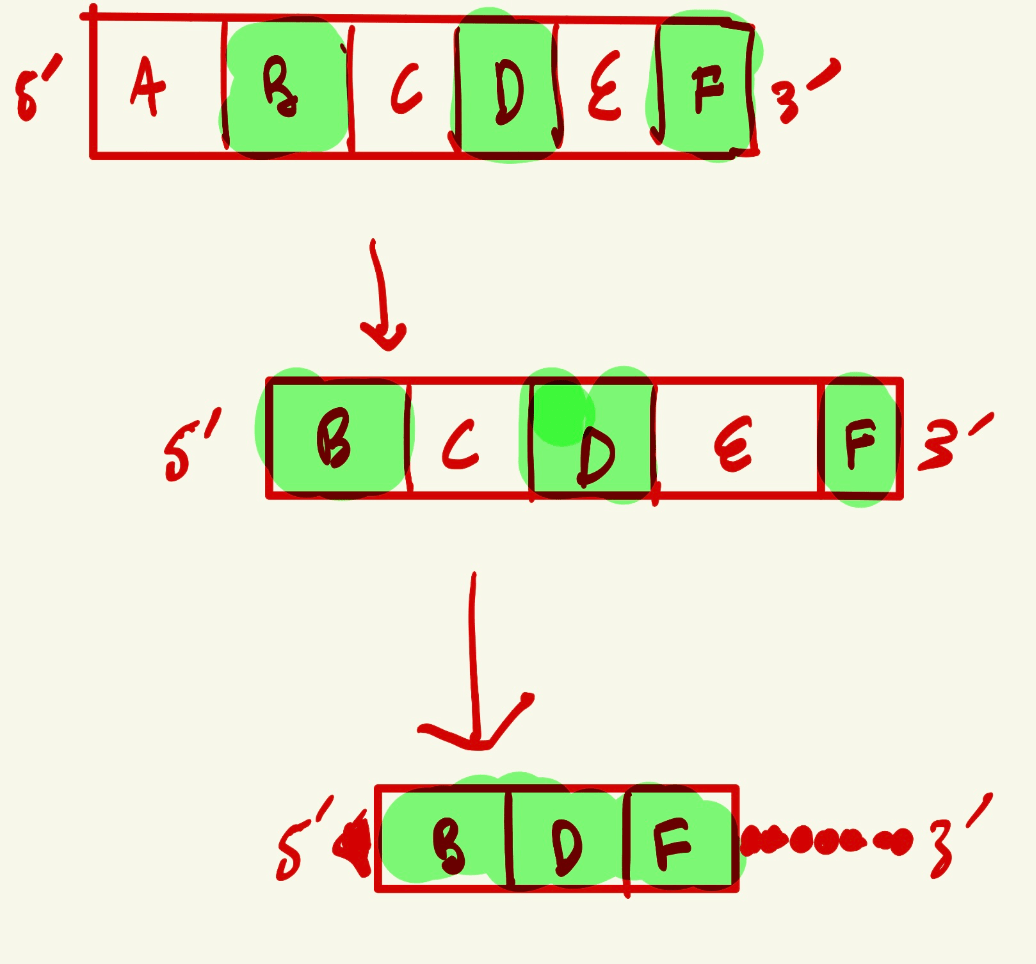
1. B D F: sections C and E were cut out and BDF were combined, implying that those are the exons
2. C and E, not A because that was the promoter, the region upstream of gene's coding sequence to initiate transcription.
3. In section A, as TATA will be in the promoter region
4. In the introns, where the GU at 5' end would and where the AG at the 3' end would be with the A branching site. These segments help intron fold with help of spliceosome, so it can be removed and the remaining exons can be joined.
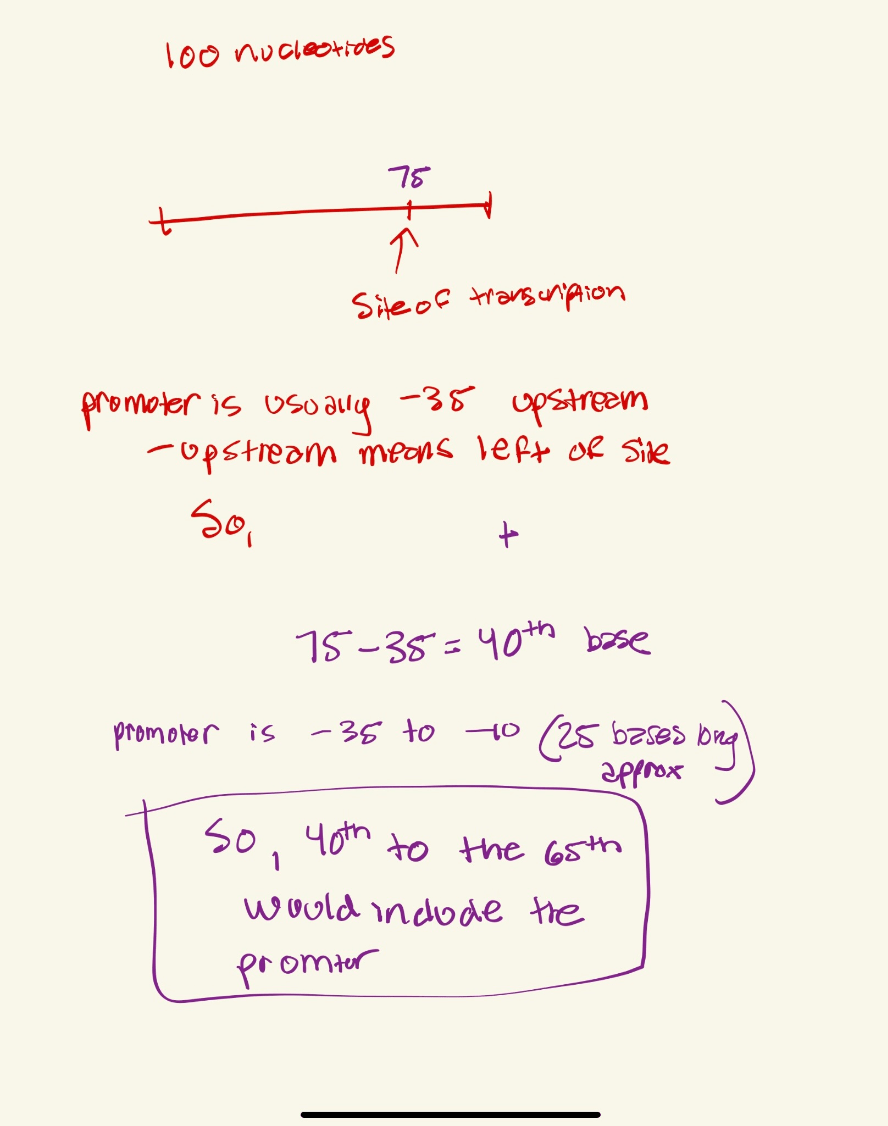
Suppose you have a 100 long nucleotide sequence of a bacteria. (5' -100 nucleotides---3')
Lets say the first site of transcription occurs at the 75th base,
Which bases would be the region of the promoter?
Write the complementary strand of 5’- GCTAG -3’ in the 5’-3’ direction. (Hint- go backwards)
5’ - CTAGC - 3’
DNA strands run antiparallel no matter what, the answer above is just reversed
5'- GCTAG -3'
3'- CGATC -5' --------->>> (written backwards) 5'- CTAGC -3'
Why are introns not present in prokaryotes
Prokaryotes are simpler, and they live in environments where rapid transcription for proteins is necessary, so intron splicing would take up too much
There is no nucleus in prokaryotes, thus there is no room for intron splicing, and the necessary splicing machine is not present for intron splicing
During elongation, RNA Polymerase II leaves the ___ at the promoter to jump to to the actual site where the first nucleotide will be synthesized
pre-initiation complex
Describe the steps of splicing
1. snRNPs called U1 and U2 (snRNPs are components of the whole spliceosome) bind to the complementary intron sequences. So, U1 binds to the "GU" at the 5' end and U2 binds to the "A" branch site.
2. Additional proteins (U4, U5, U6) are then recruited to the spliceosome
3. U1 and U4 leave spliceosome
4. the 5' end of the intron with the "GU" attaches to the "A" branch site (this disconnects the exon that was at the 5' end
5. the exon at the 5' end is connected with the exon that was at the 3' end
Give the template strand, coding strand, and non template strand of the RNA sequence 5' AGUCG 3'
Make sure to include polarity
Template strand - 3' TCAGC 5'
Coding strand - 5' AGTCG 3'
Nontemplate - 5' AGTCG 3' (Nontemplate and coding strand mean the same thing)
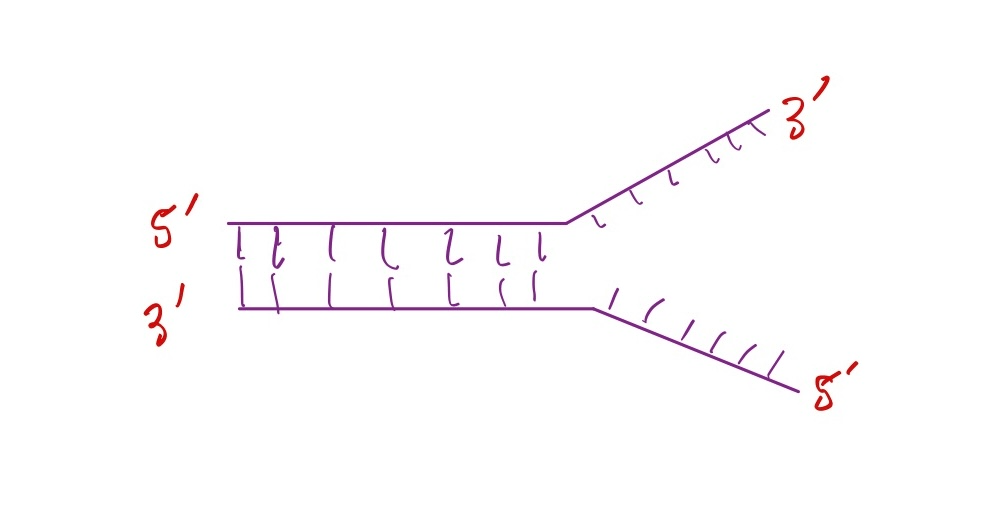
Which strand is the leading strand?
Top strand
Explanation: the direction of DNA synthesis of the top strand goes in the same direction the replication fork is opening, so it can be made continously.
How is eukaryotic transcription initiation different from prokaryotic initiation?
Prokaryotic- A sigma factor protein complex binds to the promoter of the gene which is in the -35 to -10 range, (+1 is where the actual nucleotide is transcribed), and then it recruits RNA Polymerase to come to that same site and initiate transcription (NO TATA BOX)
Eukaryotic - Within the promoter part of the gene, there is a TATA box around 25-30 bases pairs upstream of the transcription start site. TBP (TATA binding protein) binds to the TATA box. TBP is connected to the other GTFs, so TBP binding to the TATA box allows for other GTFs be attached as well. After this, RNA Polymerase II is recruited along with more GTFs to form the pre-intiation complex
What else besides the promoter is required for eukaryotic gene expression?
Enhancers
While promoters help signal to transcription machinery where to start, enhancers also control to what extent a gene should be transcribed. Enhancers can be tissue specific.
RNA processing is coordinated by the ___ of the ____
1. CTD (carboxyl -terminal domain)
2. RNA Polymerase II
Further Explanation: CTD contains the amino acids YSTSS, but 2nd and 5th Serine amino acids get phosphorylated ----->> YSPTSPS, and this phosphorylation status can recruit appropiate specific enzymes for either capping, splicing, and polyadenylation
Write the anticodon for 5' - UGA- '3
3' ACU 5'
Without single stranded DNA binding proteins, what would happen?
Single stranded binding proteins keep each individual strand of DNA straight and still apart after it has been unzipped by the helicase. Without these SSBs, the complementary bases would start reconnect with one another in the same strand, giving result to a hairpin like structure. Or the complementary bases from both separated strands could rejoin, forming a double helix again.
How is prokaryotic replication termination different from eukaryotic termination?
Prokaryotic: Factor - Independent Termination (Poly (U) tail)
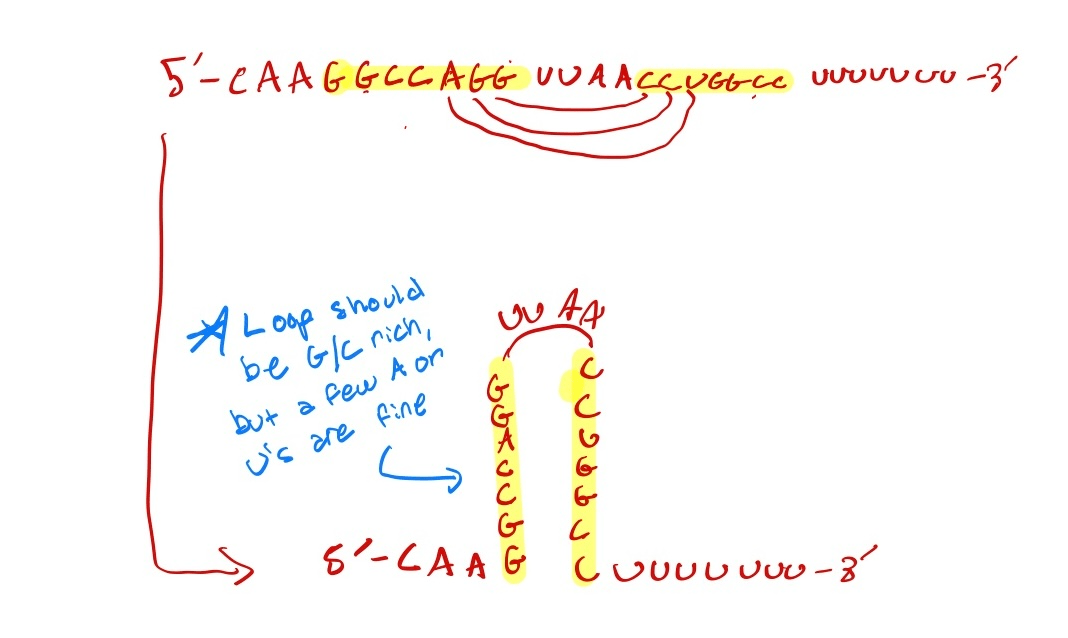 Eukaryotic Termination: Remember how part there is a Poly(A) tail that gets added to the 3' end of the RNA transcript in as a part of RNA processing/modification. The RNA polymerase encounters the Poly (A) signal that is typically 5'AAUAA '3 , this signals to the RNA Polymerase you add the Poly A tail somewhere close by. this causes the RNA Polymerase to recruit cleavage annd polyadenylation proteins. They bind to that Poly (a) signal that was 5' AAUAA, the RNA gets cleaved and polyadenylated (in other words you cut away nucleotides and replaces it with a bunch A's to make the Poly(A) tail) AS THIS HAPPENS, RNa polymerase II undergoes a change in shape that causes it to RNA transcript to detach from it, causing termination. So basically, 3' polyadenylation helps cause RNA termination
Eukaryotic Termination: Remember how part there is a Poly(A) tail that gets added to the 3' end of the RNA transcript in as a part of RNA processing/modification. The RNA polymerase encounters the Poly (A) signal that is typically 5'AAUAA '3 , this signals to the RNA Polymerase you add the Poly A tail somewhere close by. this causes the RNA Polymerase to recruit cleavage annd polyadenylation proteins. They bind to that Poly (a) signal that was 5' AAUAA, the RNA gets cleaved and polyadenylated (in other words you cut away nucleotides and replaces it with a bunch A's to make the Poly(A) tail) AS THIS HAPPENS, RNa polymerase II undergoes a change in shape that causes it to RNA transcript to detach from it, causing termination. So basically, 3' polyadenylation helps cause RNA termination
______ part of the RNA Polymerase II allows for the RNA Polymerase to exit from the pre-intitation complex to start transcribing
This part also serves as the binding site for the enzymes necessary for the capping, splicing, and polyadenylation.
carboxy terminal domain
The CTD amino acids can get phosphorylated, so this phosphorylation status controls which proteins can bind or whether RNA polymerase can exit to go start synthesis
Describe in the 3 ways RNA is processed, where it happens and at what point in time during transcription
1. 5' capping - a modified nucleotide,7-methylguanosine, called the 5' cap is added as the RNA strand emerges as RNA polymerase is actively transcribing. This happens while in the nucleus. Purpose of this is so that the 5' end of RNA is protected and not degraded as it is transported to the cytoplasm. This 5' cap also allows for proteins involved in transport to bind. Occurs in nucleus. Happens early in transcription
2. 3' Polyadenylation - RNA Polymerase recognizes a sequence (5'-AAUAAA -3') and this is its signal to that the polyadenylation modification must occur around there. So upon that signal, the polyadenylation proteins are recruited by the CTD of the RNA polymerase II. These proteins then cut the RNA 15-30 nucleotides, and add in 50-250 A's to the 3' end. This is called the Poly- A tail and it serves to protect 3' from degradation. Occurs in nucleus as well. Happens end of transcription
3. Splicing - cuts away introns, and joins exons. The exons are the transcribed regions that actually are going to be used to code for a protein. Introns are transcribed junk we do not need. Occurs in nucleus. Can happen after transcription or as it is occuring.
Here is the template DNA strand, write out the polypeptide chain that would result, make sure to label the amino group and the carboxyl group on appropriate side
3' TACCTGTAA 5'
1. First, transcribe into RNA strand, this is the template so, write out complement
5' AUG GAC AUU 3'
2. Write the appropriate amino acid based off of what each codon codes for
NH3+ - Met-Asp-lle -COO-
Which strand is the template strand for the RNA sequence 5’- UUGGAC -3’
5’- AACCTG -3’
5’- GTCCAA -3’
3’- TTCCTG -3’
3’- GCGAC -3’
None of the above
B
1. Write out the template strand
2. Reversing the sequence formed by step above results in the answer B