Give the polypeptide for this mRNA transcript
3' AGU UUU GUA '5
Reverse the sequence:
5′ - AUG UUU ACU - 3′
NH3+ - Met - Phe - Thr - COOH-
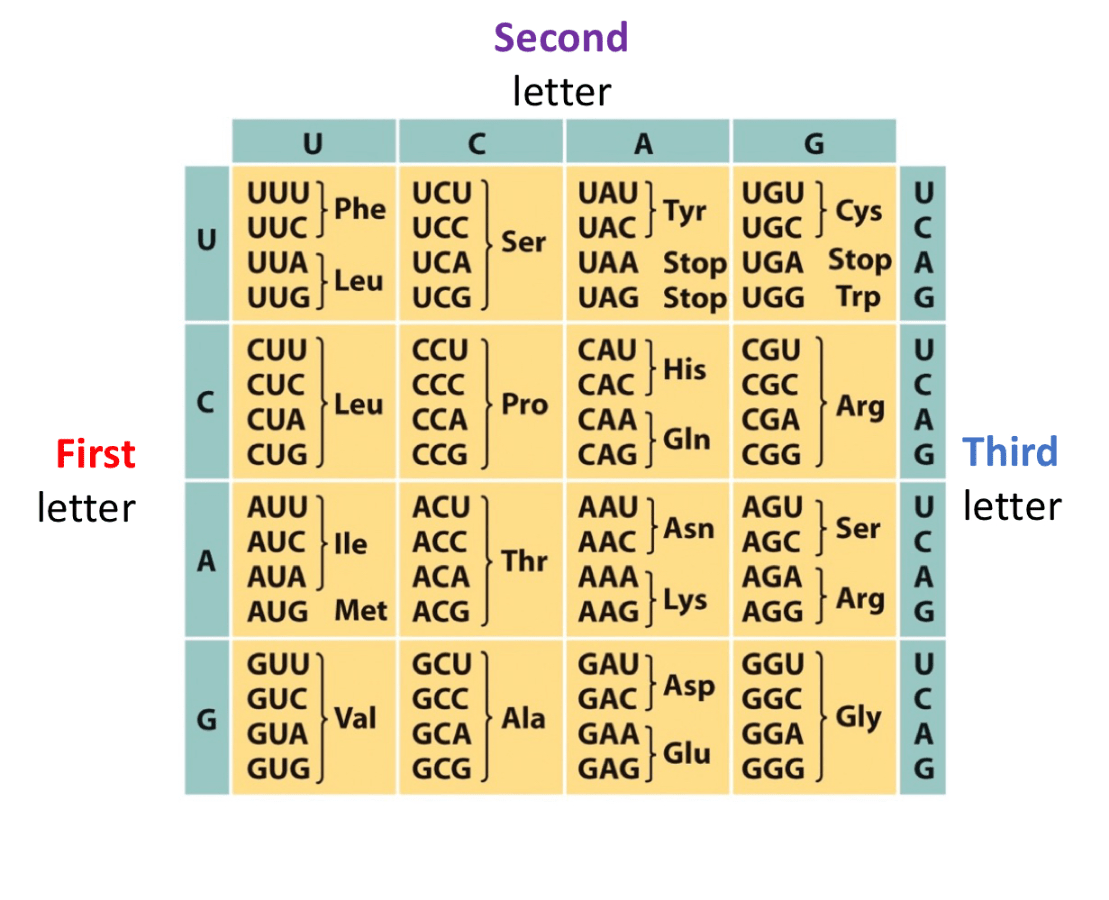
What are the tRNA anticodons for the amino acid, Proline? Make sure polarity is included
Proline has 3 codons that code for it
5' CCU '3 --->>> 3' GGA '5
5' CCC '3 --->>> 3' GGG '5
5' CCA '3 --->>> 3' GGU '5
5' CCG '3 --->>> 3' GGC '5
*** Anticodons run in opposite direction and are complementary.
What is tRNA?
This molecule carries amino acids to the ribosome and matches its anticodon with the mRNA codon during translation.
What is gene expression regulation
This process turns genes on or off depending on the cell’s needs.
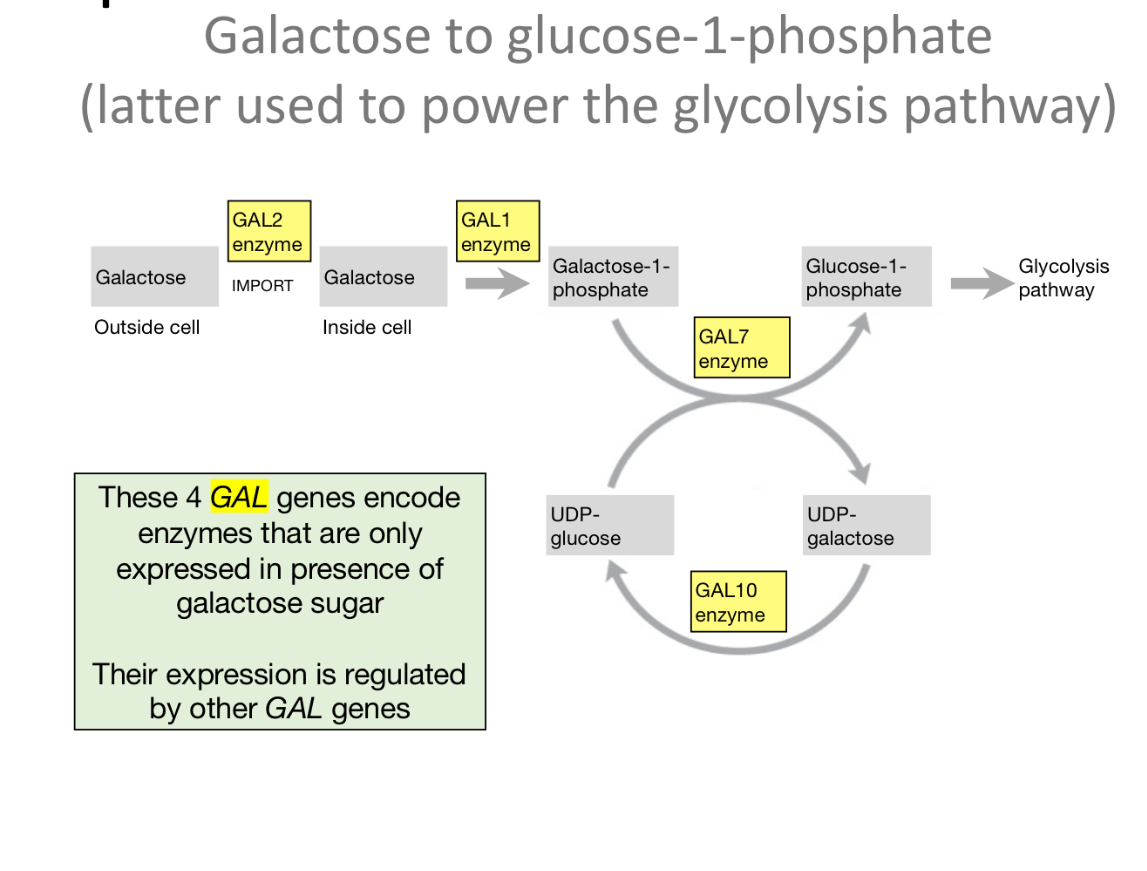
What are Gal Enzyme genes?
Gal enzyme genes are genes that encode enzymes involved in the galactose metabolism pathway
In eukaryotic translation initiation, what does the small ribosomal subunit first bind to?
A. The start codon AUG directly.
B. The 3’ poly-A tail of mRNA.
C. The 5’ cap of mRNA.
D. The promoter region of DNA.
C
If a mutation caused the removal of the 5’ cap on an mRNA, what is the most likely outcome?
A. Translation initiation would fail due to ribosome binding issues.
B. Translation would start at a random codon downstream.
C. The mRNA would still be translated normally.
D. The protein produced would be longer than normal.
A
The 5’ cap is crucial for ribosome recognition and binding in eukaryotic translation initiation.Without the cap, the small ribosomal subunit and initiation factors cannot properly bind to the mRNA, so translation initiation is blocked or severely reduced.This results in little to no protein being made from that mRNA.
Why is the genetic degenerate?
This degeneracy offers protection against mutations, as a change in one or more bases of a codon might not change the encoded amino acid, and it enhances translation efficiency by allowing fewer tRNA molecules to recognize all codons. The reason the genetic code is degenerate because there are 64 possible three nucleotide codons, but only 20 amino acids present
Why does gene expression need to be controlled? 3 reasons
1. We need to make sure proper proteins at are made at right time and place not all the time. This allows proper development, specialized cells have the right properties, etc
2. Expression of some genes can cause harm if expressed at wrong time. For example, you would not want cell division genes constantly turned one, that leads to uncontrolled cell growth, potentially causing cancer
3. Making unnecessary protein if it isn't even needed is a waste of energy.
The _____ TF protein is required to activate the GAL enzyme genes.
Gal 4
It binds to an upstream activation sequence (UAS), this is a sequence located upstream of the GAL enzyme genes
The genetic code has 64 codons, with 3 being stop codons. Which of the following statements is TRUE?
A. Each amino acid is matched to exactly one tRNA molecule.
B. Some tRNAs can recognize more than one codon due to wobble base pairing.
C. All amino acids are encoded by exactly one codon.
D. There are 64 different tRNAs, one for each codon.
B
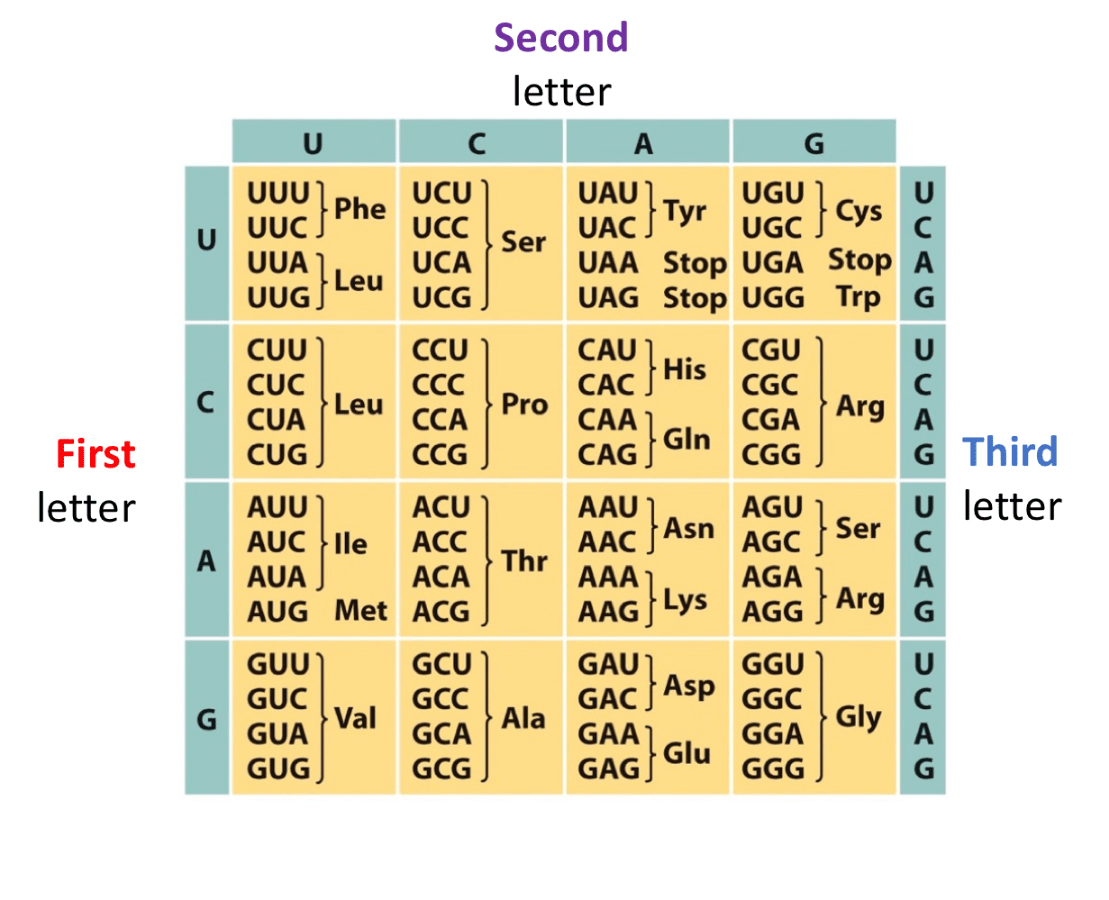
A tRNA with anticodon 3' UCU '5 pairs with which codon(s)? Show the codon(s) it pairs with and consider wobble base pairing.
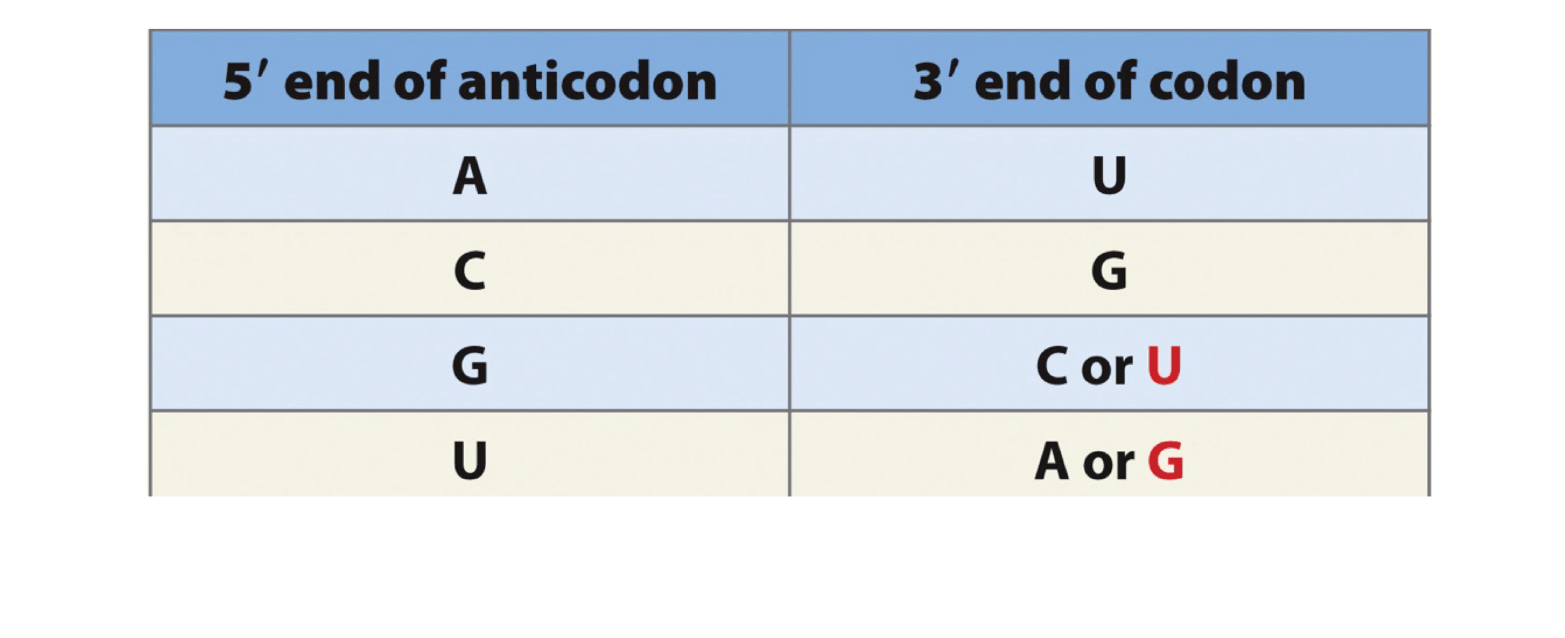
5' AGA '3 (Standard)
5' AGG '3 (Non Standard)
**Only scenario where standard rules are broken, however you cannot do U and C bonded because that pair is too unstable
How do you do initiate translation in prokaryotes vs eukaryotes
We need to position the first tRNA (AUG the start codon) at the P-site of the ribosome
Prokaryotes: There is a shine delgarno sequence called 5' AGGAGGU '3 5-6 bases upstream of the first AUG, SO find the shne delgarno sequence and then after that find the first AUG codon (you will have mulitple AUG codons in the sequence but choose the first one after the shine- delgarno) Overall, this shine dalgarno sequence serves as a ribosomal binding site, where it binds to a complementary sequence somewhere on the ribosome, this aligment allows for the AUG codon to be in P-site
Eukaryotes: small ribosome subunit binds to 5'cap of mRNA, and then reads through it from that point till it hits its first AUG. Initiation codon is found within the Kozak sequence 5' CC(A/G)CCAUGG-3'
Describe the levels at which gene regulation occur.
1. Transcriptional: We have can have extra regulatory proteins bind to sections upstream of promoter to enhance transcription, or we can modify chromatin structure
2. Translation: We can control which mRNA are translated
3. RNA processing: Alternative splicing gets rid of unnecessary mRNA
4. Mature mRNA regulation: Regulation of mRNA transport to cytoplasm
5. Post translational: Modifying AAs to alter their functionality or regulating protein stability.
How does Gal4 protein turn gene expression off or on?
***** Keep in mind this is only to transcribe the genes that code for the enzymes for the metabolism of galactose, every gene differs in it's own way in how their own TFs act, this is just a general example.
Gal4 requires its activation domain to initiate transcription so
Without that activation domain, you cannot express the gene
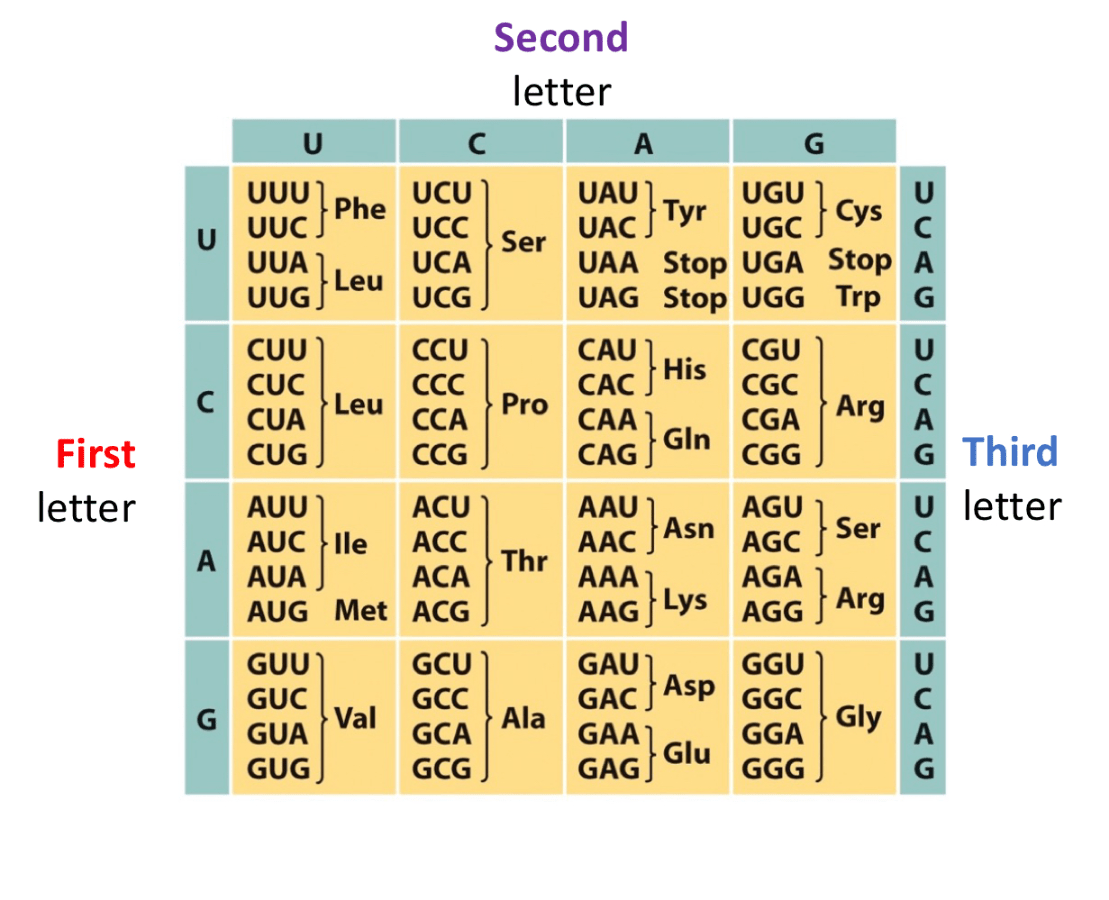
This polypeptide chain is changed due to a mutation caused by a single nucleotide substitution, where the base at position __ of the mRNA was altered.
NH3 - Met - Ala - Gly - Lys - COO-
to
NH3 - Met - Thr - Gly - Lys - COO-
The codons for Ala
GCU GCC GCA GCG
For Thr,
ACU ACC ACA ACG
These two sections of codons differ only in their first letter, so mutation occurred at base 4 of mRNA
Down below is a segment of double stranded DNA, write out the polypeptide chain to result if top strand is template.
5' CCG GAT CAT ATA '3
3' GGC CTA GTA TAT '5
Top strand is template so complement mRNA would be
3' GGC CUA GUA UAU '5
For convenience, reverse it because mRNA is translated from 5' to 3
5' UAU AUG AUC CGG '3
N- Met-Lle- Arg -C
*** Remember that first amino acid will always be the AUG codon
What happens in the three binding sites for tRNAs?
1. A site, also called aminoacyl tRNA binding site
This is the section where charged tRNAs carrying the amino acid come, they use their anticodon to attach to the complementary codon in the mRNA within the A site
2. P or peptidyl site
The amino acid of the growing chain in P site disconnects from that tRNA and connects to the amino acid the tRNA in site A bought
3. The uncharged tRNA leftover from P site goes to the E site, where it exits from the ribosome
Describe the function of each
Core promoter
Proximal enhancer
Distal enhancer
Core promoter: signals transcription machinery where to start synthesizing RNA
Proximal enhancer: Occurs right close upstream of promoter sequence, binds TFs . Binds TFs so that level of transcription can be regulated
Distal enhancer: Bind TFs as well, and controls when and how much of a gene to produce according to what cell type it is.
What does Gal80 do?
binds to Gal4 activation domain, prevents other proteins from binding to Gal4 such as transcription machinery to initiate transcription. . So Gal80 acts as a repressor of GAL enzyme gene expression
2 Parts to this Question
1. Below is a double strand of DNA, design the polypeptide chain if it is transcribed left to right. (left to right is your hint of which strand will be template vs coding
3' TAC GCC AGT CCT '5
5' ATG CGG TCA GGA '3
2. In the DNA double strand above, the C/G at the 10th base is replaced with A/T, respectively, now write the new resulting polypeptide.
1. If it is going left to right, the template strand has to be the top one, only way mRNA be created 5' to 3'
Transcribe
5' AUG CGG UCA GGA '3
Translate
NH3+ - Met- Arg - Ser- Gly - COOH
2. We change the 10th base position to A/T so,
3' TAC GCC AGT ACT '5
5' ATG CGG TCA TGA '3
Top strand is still template, so transcribe new mRNA
5' AUG CGG UCA UGA '3
Translate
NH3+ - Met- Arg - SER - COOH
That mutation turned the last codon to a stop codon, no need to write STOP
1. Insertion/ deletion: Frameshift, effect severe
2. Substitution that causes normal codon to be changed to stop codon, so this causes a early stop codon, effect severe as causes a prematurely stopped polypeptide
3. Substitution that changes one Amino acid to another, effect not as bad as the first two
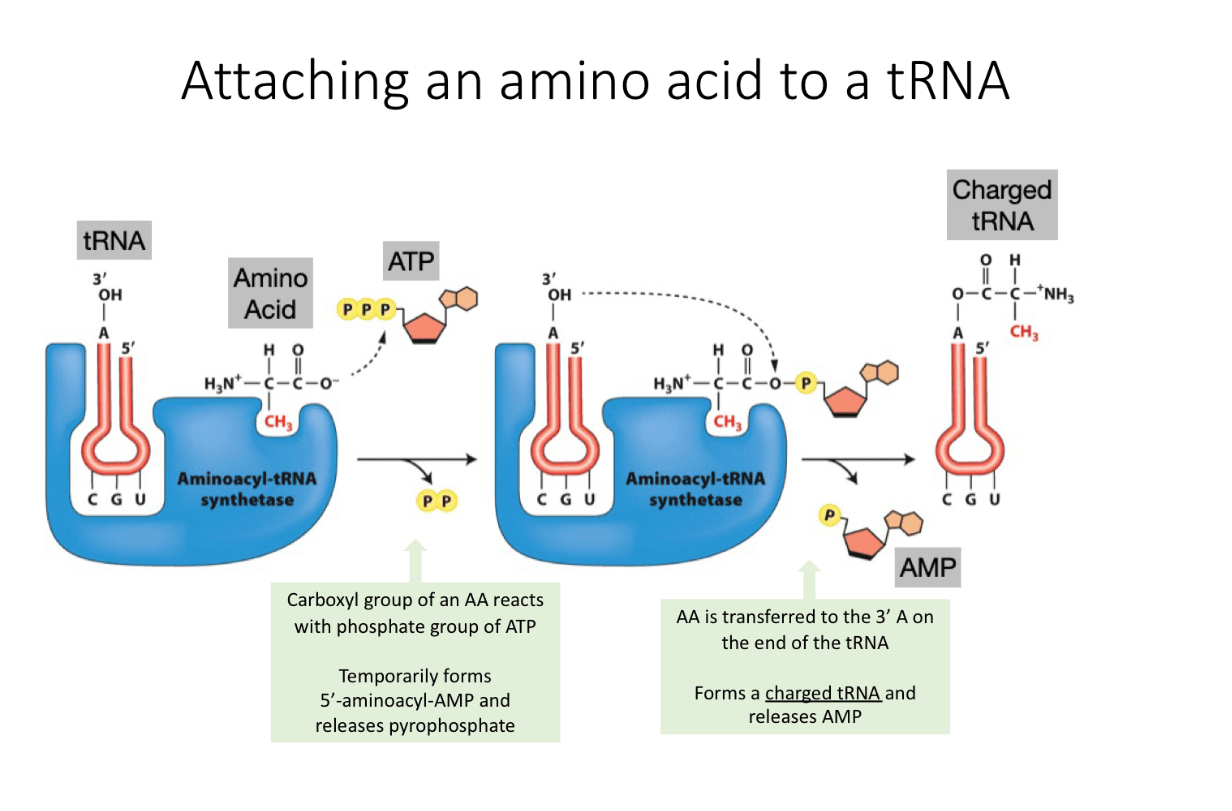
Describe the steps of attaching an amino acid to a tRNA
These are the critical proteins for transcription and also aid in proper gene regulation give the functions of each
RNA Polymerase II and GTFs:
Transcription factors (TFs):
Transcriptional Coregulators:
RNA Polymerase II and GTFs: come to the core main promoter and initiate transcription
Transcription factors (TFs): Bind to the proximal and distal enhancers. Different set of TFs for expression of different genes
Transcriptional Coregulators: Can activate or repress transcription. Do not bind directly to DNA, they rather do it indirectly like like binding to TFs, and controlling if they can do their job
What does gal3 in presence of galactose do?
What does gal3 in absence of galactose do?
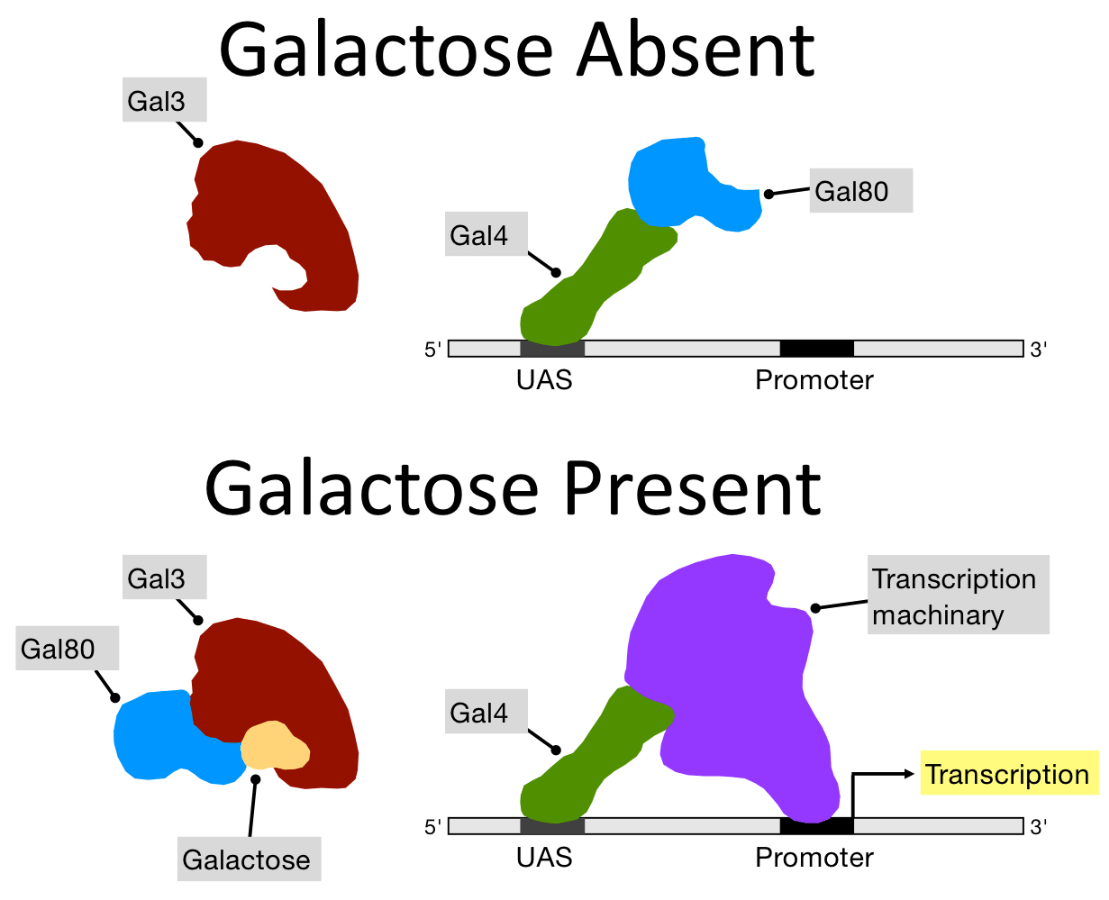
***The general role of Gal3 is to release gal80 from gal4, allowing transcriptional activation
When galactose is present it binds to gal3, WHICH causes gal3 to have a shape that can bind to gal80, taking gal80 away from gal4, so gal4 can do its job of activating transcription
When galactose is absent, gal 3 has a specific shape that doesn't bind to gal80, causing gal80 to remain stuck to gal4, inhibiting transcription activation of the genes.