How are phosphodiester bonds formed?
3’ hydroxyl (OH) on last nucleotide attacks 5’ phosphate on dNTP => releasing phosphates and forming a phosphodiester linkage
What is a base tautomer and what do they result in?
What is telomerase and what purpose does it serve?
Base tautomers are the result of a proton shift in a base. Base tautomers pair with non-standard base partner => mismatch pairing
Telomerase = A reverse transcriptase that synthesizes DNA using an RNA template. It extends ends of DNA (telomeres) following replications to avoid shortening of chromosomes
List three examples of when recombination occurs between homologous sequences,
Also difference between Tyrosine and Serine?
1. In response to DNA damage
i. Repair of DNA double-stranded break
2. Spontaneously during meiosis
3. When catalyzed by enzymes
i. tyrosine and serine recombinases (DNA strand breaks as 2 sequential single VS 2 simultaneous double)
Explain two ways microRNAs can silence genes.
Precise pairing: results in cleavage of the target RNA
Imprecise pairing: represses translation of the target RNA
Explain the difference between silent, missense, and nonsense mutations.
BONUS: What is the tRNA attached to when it occupies the A, P, and E sites of the ribosome?
a. Silent – change the mRNA sequence to a synonymous codon (doesn’t change polypeptide sequence)
b. Missense – change the mRNA sequence to a codon for a different amino acid (changes polypeptide sequence)
c. Nonsense – change the mRNA sequence to a stop codon (truncated polypeptide)
BONUS:
A – carboxy terminus of incoming amino acid (aminoacyl tRNA)
P – carboxy terminus of polypeptide chain
E – no longer attached to an amino acid; ready for exit
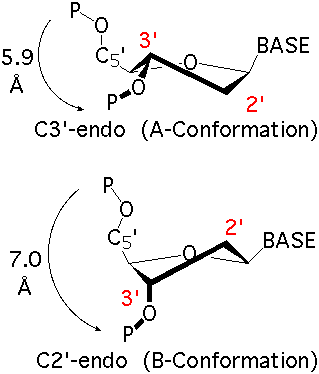
Explain what a "sugar pucker" means.
The pentose ring of a nucleotide can take five different conformations
The conformation affects distance between phosphates => affecting structure of the polynucleotide chain
List three characteristics of DNA polymerase
a. Requires a primer to initiate synthesis
b. Uses phosphoryl-transfer reaction to add dNTPs to 3’ end of last nucleotide in the polynucleotide strand (works 5’ to 3’)
c. Active site contains Asp which coordinate Mg+2
i. Activates 3’OH attack on dNTP phosphate
ii. Stabilizes phosphates negative charge
d. Has 3’-to-5’ exonuclease activity that proofreads new DNA strand
What are transposons, what purpose do they serve, and what is one example?
a. Mobile DNA elements that move around genomes
b. Cause genetic changes à genetic diversity
c. Antibody or phenotypic diversity, can cause mutations that result in disease
Explain how transcription factors interact with DNA and 3 ways they can inhibit gene expression
Interaction
a. Amino acid side change form hydrogen bonds with nucleotide bases
b. Most proteins bind in major groove
c. Proteins have polypeptide motifs that recognize specific nucleotide sequences
Gene expression inhibition
a. Bind DNA at a repression domain
b. Interfere with an activator protein (block binding site or activity)
c. Recruit corepressors
When sequencing DNA how can you determine which nucleotide is in the terminal position?
Each ddNTP has a different fluorescent label attached
List three things that affect the melting temperature of DNA, and in what direction.
1. Non polar solvents – disrupts stacking => destabilizes DNA (lower Tm)
2. Cations – shield phosphate charges => stabilize DNA (higher Tm)
3. pH – affects hydrogen bonding => very high or low destabilizes DNA (lower Tm)
4. Base pair mismatches – destabilize double-stranded DNA (lower Tm)
List four types of DNA mutations caused by replications errors
1. Transitions: purine-to-purine, or pyrimidine-to-pyrimidine change
2. Transversions: purine-to-pyrimidine or pyrimidine-to-purine change
3. Small deletions or insertions: may cause a codon “frame shift”
4. Large deletions or insertions, inversions, or chromosome fusions
Explain the difference (including orientation) between the coding and template strands of DNA.
Coding strand (5’ => 3’) will be identical to the mRNA sequence (except: TàU)
Template strand (3’ => 5’) will be complementary to the mRNA sequence
What causes DNA methylation and what is the result of the methylation?
What are the three main types of histone tail modifications? Give an example of one modification that can increase gene expression.
DNA methylation is caused by environmental exposures (or inherited). It silences genes by affecting chromatin status and preventing binding of regulatory factors.
Histone tail modification: Phosphorylation, Methylation, & Acetylation
-Acetylation of lysine residues in histone tail disrupts electrostatic interactions between histone and DNA making genes more accessible for transcription.
Where is the Shine-Dalgarno sequence located and what purpose does it serve?
a. Located in the 5’UTR of prokaryotic mRNA
b. Allows translation initiation through binding of 16S rRNA in 30S ribosome subunit
c. Helps distinguish the initiating AUG start codon from other AUG codons
Explain what histones are and how they are modified to regulate gene expression.
Histones= proteins which DNA is wrapped around around to form nucelosomes
-tightly package DNA into nucleus (gene inaccessible: off)
Histones are acetylated to allow gene transcription
-DNA unwound from histone allowing transcription factors to reach DNA (gene accessible: on)
List four types of DNA mutations caused by chemical changes to bases in DNA
a. Base alkylation: disrupts base pair hydrogen bonding => mismatch
i. ie: O6-methylguanine substitutes for adenosine
ii. Corrected by direct repair involving methyltransferases
b. Deamination: loss of exocyclic amino groups (thymine cannot be deaminated)
i. ie: cytosine to uracil
ii. Corrected by base-excision repair using DNA glycosylases
c. Depurination: N-glycosidic bond between base and pentose cleavedà purine base removed and left with apurinic residue
i. Corrected by base-excision repair
d. Pyrimidine dimer – UV light causes bond to form between adjacent T (or C) nucleotides à kinks DNA and disrupts hydrogen bonding
i. Corrected by direct repair (or nucleotide excision repair) using DNA photolyases
Explain how RNA polymerase differs from DNA polymerase (template, initiation, exonuclease, product)
a. Both use DNA as a template
b. RNA polymerase recognizes a promoter sequence rather than a primer
c. RNA polymerase does not have exonuclease activity (no proofreading)
d. The product of RNA polymerase is RNA rather than DNA.
List three ways mRNA is processed and why these modifications are important.
What is RnaseP and how does it function?
1. Introns are spliced out allowing for different protein products from splice variants
2. 7-methyl guanosine cap is added which aids in translation and protects the 5’ end from degradation
3. 3’ polyA tail is added which signals nuclear export, stabilizes mRNA, and promotes translation.
RnaseP is a ribonuecleoprotein complex with a catalytic RNA subunit that cuts transcripts at precise locations to generate ribosomal and transfer RNAs
Explain the three main steps in polymerase chain reaction (PCR)
Denaturation – temperature increased to separate DNA strands
Annealing – temperature decreased to allow primers to base pair to complementary DNA template
Extension – Polymerase extends primer (5’ à 3’) to form nascent DNA strand
*DNA primer is included in final PCR product*
What is the purpose of topoisomerase and how does it work?
Purpose - relieve strain in DNA by breaking and rejoining strands
Mechanism (pg. 20-21 in notes)
i. Enzyme in closed conformation: 3’OH on tyrosine of topoisomerase initiates nucleophilic attack on 5’ phosphate in DNA strand to break DNA (phosphoryl-transfer reaction).
ii. Enzyme changes to open conformation: moving unbroken DNA strand through break in first strand
iii. Enzyme in closed conformation: liberated 3’OH attacked 5’ phosphotyrosyl protein-DNA linkage (DNA bound to Tyr) to relegate cleaved DNA strand

List four different repair processes and a brief description of how they work.
1. DNA polymerase: 3’-to-5’ exonuclease activity
i. Mismatch removed right away and reaplaced with correct nucleotide
2. Methyl-directed mismatch repair: repairs mismatches made by DNA polymerase
i. Recognizes mistakes in unmethylated daughter strand, nucleases break new strand, DNA polymerase replaces area removed and ligase seals gap
3. Base excision repair: Repairs deaminated and depurinated nucleotides damaged by chemicals.
i. Glycosylase cleaves bond between base and sugar *if the base is there*
ii. Nucleases cut the damaged strand, DNA pol I fills in gap and ligase seals
4. Nucleotide excision repair: Repairs pyrimidine-pyrimidine dimers
i. Similar to base excision – nucleases cut damaged strand, DNA pol I fills in gap and ligase seals
Explain the two ways transcription is terminated in prokaryotes.
a. Rho-dependent – Rho site exposed à Rho helicase melts DNA-RNA hybrid
b. Intrinsic – RNA hairpin forms followed by string of UUU’s
Explain how the spliceosome and self-splicing introns are similar and different
a. Spliceosome – uses small ribonucleoproteins (snRNPs), 2’ OH of adenosine within 5’ end of intron initiates 1st nucleophilic attack, 3’ OH of the 5’ exon initiates 2nd nucleophilic attack.
b. Type I self-slicing – No proteins involved, 3’ OH of guanosine from elsewhere in the transcript initiates 1st nucleophilic attack, 3’ OH of the 5’ exon initiates 2nd nucleophilic attack.
c. Type II self-splicing – No proteins involved, 2’ OH of adenosine in the intron initiates 1st nucleophilic attack, 3’ OH of the 5’ exon initiates 2nd nucleophilic attack.
Explain the three main steps in CRISPR-Cas9 technology
a. Guide RNAs (gRNAs) bind to target genomic sequence
b. Cas9 endonuclease cuts the DNA
c. Double-stranded breaks are repaired by homologous recombination or non-homologous end joining (NHEJ)