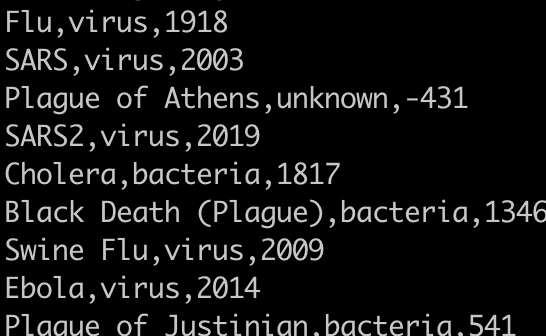
How many instances of the flu are there?
grep -c "flu" pandemics.csv
This will give you a count of zero because grep is case sensitive. You need to either:
1) use the -i flag to ignore case or
2) capitalize the first f in flu.
How many movies are there?
wc -l some_pandemic_movies.txt
cut -f 1 some_pandemic_movies.txt | wc -l
awk -F "\t" some_pandemic_movies.txt | wc -l
How many movies were released in 2023?
grep -c 2023 some_pandemic_movies.txt
grep 2023 some_pandemic_movies.txt|wc -l
How many sequences are there in the file?
grep -c '>' flu_sequences.fa
grep '>' flu_sequences.fa | wc -l
Log onto logrus
ssh agomez@gateway.training.ncgr.org
This is missing the port (44111).
ssh -p 44111 agomez@gateway.training.ncgr.org
Print only the year (column 2).
cut -f 2 some_pandemic_movies.txt
awk '{print $2}' some_pandemic_movies.txt
awk -F "\t" '{print $2}' some_pandemic_movies.txt
Sort the file by year (second column)
sort -nk2 some_pandemic_movies.txt
sort -k2 some_pandemic_movies.txt (because all the years are 4 numbers, this will give you the same answer but best practice is to use -n regardless).
What is the earliest (field 6) flu sequence in the file? Make sure you remove blank lines.
grep '>' influenzaA_sequences.fa | awk -F'|' '{print $6}'|grep -v '^$'|sort -n|head -1
grep '>' influenzaA_sequences.fa | cut -f 6 -d '|' |grep -v '^$'|sort -n|head -1
etc
Look at the top part of a file that you loaded into the variable dns in R.
head dns
That is linux syntax. In R you have to use parentheses:
1) head(dns)
If you want to see 10 lines
2) head(dns,n=10) or head(dns,n=10L)
Grab all lines ith pandemic movies that were made before 1980 (column 2)
awk '$2<1980{print}'
awk '$2<=1979{print}'
awk -F "\t" '$2<1980{print}'
awk -F "\t" '$2<=1979{print}'
How many unique movie titles are there?
cut -f 1 some_pandemic_movies.txt | sort -u | wc -l
cut -f 1 some_pandemic_movies.txt | sort | uniq | wc -l
awk -F "\t" some_pandemic_movies.txt | sort -u | wc -l
etc
How many countries (field 5) are represented in this file? Note that some samples will have a blank entry in the country field and shouldn't be counted.
grep '>' influenzaA_sequences.fa | awk -F'|' '{print $5}'|sort -u| grep -v '^$' | wc -l
grep '>' influenzaA_sequences.fa | cut -f 5 -d '|' |sort -u| grep -v '^$' | wc -l
etc
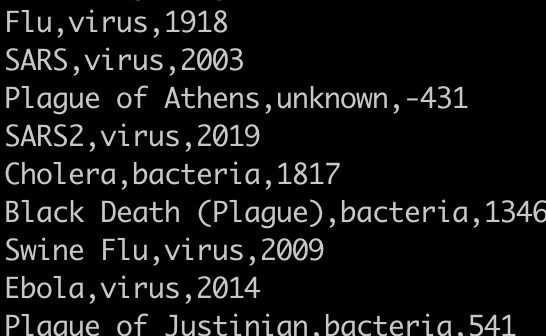
Print the pathogen column
awk '{print $2}' pandemics.csv
1) You need to change the delimiter to a comma.
2) Alternatively, you could use:
cut -d, -f2 pandemics.csv
Print only the movie column (1st column).
cut -f 1 some_pandemic_movies.txt
awk -F "\t" '{print $1}' some_pandemic_movies.txt
How many of each pathogen (column 3) are there?
cut -f 3 some_pandemic_movies.txt | sort | uniq -c
cut -f 3 some_pandemic_movies.txt | sort -u | wc -l
awk -F "\t" '{print $3}' some_pandemic_movies.txt | sort | uniq -c
etc
How many H1N1 sequences from 2009 are there?
grep H1N1 influenzaA_sequences.fa | grep 2009
grep '>' influenzaA_sequences.fa | grep H1N1 | awk -F '|' '$6~/2009/{print}'|wc -l
Note that there are 8 that have an empty collection date but say 2009 elsewhere in the header so you will get two different counts.
etc
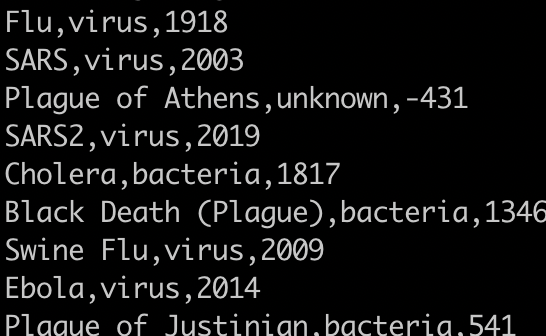
Find the unique pathogens.
awk -F, '{print $2}' sort.pandemics.csv | uniq
You need to sort it first. If you run it like this it will only collapse duplicates that are on adjacent rows. You will get: virus, unknown, virus, bacteria, virus, bacteria.
1) awk -F, '{print $2}' sort.pandemics.csv | sort | uniq
1) awk -F, '{print $2}' sort.pandemics.csv | sort -u
Grab all lines with pandemic movies that were made between 1980 and 2000 (column 2).
awk '$2>=1980&&<=2000{print}'
awk '$2>1979&&<2001{print}'
awk -F "\t" '$2>=1980&&<=2000{print}'
etc
How many movies start with "Virus"?
grep -c "^Virus" some_pandemic_movies.txt
cut -f 1 some_pandemic_movies.txt | grep -c "^Virus$"
awk -F 1 some_pandemic_movies.txt | grep -c "^Virus$"
Figure out how many of each host (field 7) there are and sort them from the host with the fewest to the most sequences.
grep '>' influenzaA_sequences.fa | cut -f 7 -d'|' | sort |uniq -c |sort -n
grep '>' influenzaA_sequences.fa | awk -F'|' '{print $7}' | sort |uniq -c |sort -n
etc